Coordinate partitioning
Partitionning of the generalized coordinates into independent and dependent variables (for constrained multibody systems only).
For more information, see Robotran theoritical framework.
- Note
- This module must be run once prior to any other module in case of multibody model containing loop and/or user-constraint.
Code sample
For running the coordinate partitioning, insert the following code in your main script and adapt it to your need :
%% 2. Coordinate partitioning [
mbs_exe_part] % For constrained MBS only
%--------------------------------------------------------------------------
MBS_user.process = 'part';
qu_id = [id_joint1, id_joint4, id_joint6];
mbs_data =
mbs_set_qu(mbs_data,qu_id); % Set variables [qu_id] as independent
qv_id = [id_joint3, id_joint5];
mbs_data =
mbs_set_qv(mbs_data,qv_id); % Set variables [qv_id] as dependent
mbs_data = mbs_set_qc(mbs_data,qc_id); % Set variables [qc_id] as
driven
opt.part = {
'rowperm',
'yes',
'threshold',1e-9,
'verbose',
'yes'};
% other options : 'visualize', 'clearmbsglobal' % Help about options on www.robotran.be
[mbs_part,mbs_data] =
mbs_exe_part(mbs_data,
opt.part); % Coordinate partitioning process
% Coordinate partitioning results
disp('Coordinate partitioning results');
disp(['Sorted independent variables = ', mat2str(mbs_part.ind_u)]);
disp(['Permutated dependent variables = ', mat2str(mbs_part.ind_v)]);
disp(['Permutated independent constraints = ', mat2str(mbs_part.hu)]);
disp(['Redundant constraints = ', mat2str(mbs_part.hv)]);
Use
The partitioning is performed by the function mbs_exe_part().
The user specifies the desired options of the partitioning in MBS_part_opt structure. The results are provided in the MBS_part structure.
The function calls successively :
See diagram below for more details :
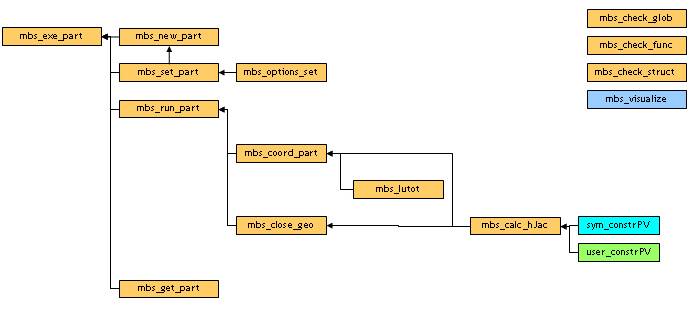
partioning function diagram

 1.8.17
1.8.17